Example script illustrating the stimuli
Contents
Load in the stimuli
load('stimuli.mat');
whos
Name Size Bytes Class Attributes
bpfilter 21x21 3528 double
conimages 1x260 247497712 cell
images 1x260 1664909120 cell
Inspect the stimuli
figure;
imagesc(double(images{12}(:,:,1)));
axis equal tight;
colorbar;
title('Stimulus frame');

whframe = 1;
res = 256;
mkdir('thumbnails');
for p=1:length(images)
imwrite(uint8(imresize(double(images{p}(:,:,whframe)),[res res])), ...
sprintf('thumbnails/thumbnails%03d.png',p));
end
Warning: Directory already exists.
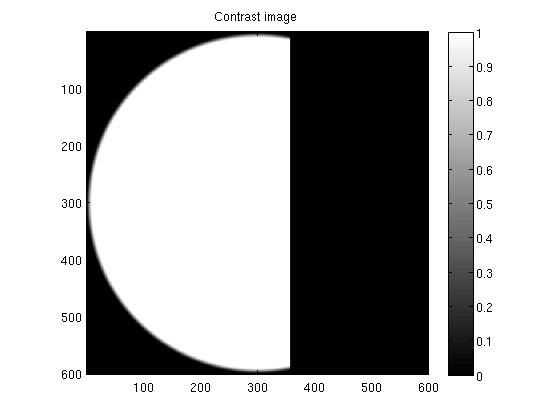
Inspect the contrast images
figure;
imagesc(double(conimages{12}));
axis equal tight;
colorbar;
title('Contrast image');
res = 256;
mkdir('conimages');
for p=1:length(conimages)
if ~isempty(conimages{p})
imwrite(uint8(255*imresize(conimages{p},[res res])), ...
sprintf('conimages/conimages%03d.png',p));
end
end
Warning: Directory already exists.
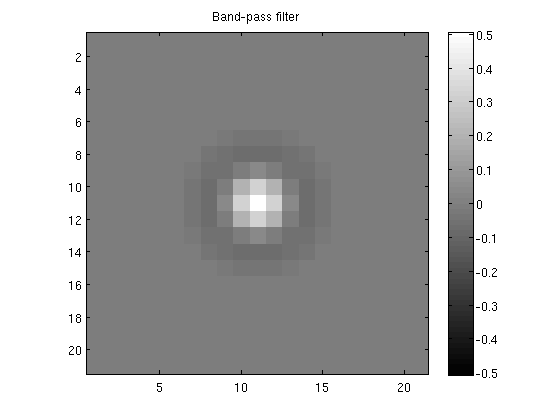
Inspect the band-pass filter
mx = max(abs(bpfilter(:)));
figure;
imagesc(bpfilter,[-mx mx]);
axis equal tight;
colorbar;
title('Band-pass filter');
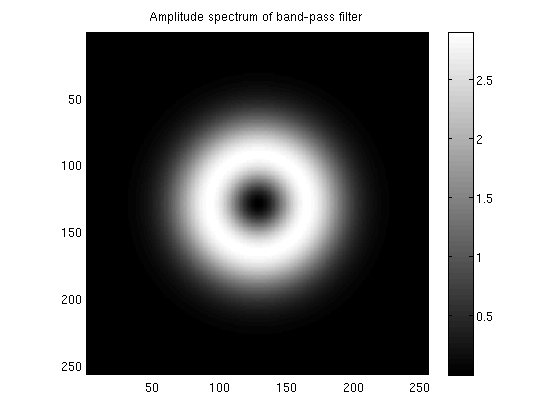
res = 256;
fov = 12.7;
temp = zeros(res,res);
temp(1:size(bpfilter,1),1:size(bpfilter,2)) = bpfilter;
temp = fftshift(abs(fft2(temp)));
figure;
imagesc(temp);
axis equal tight;
colorbar;
title('Amplitude spectrum of band-pass filter');
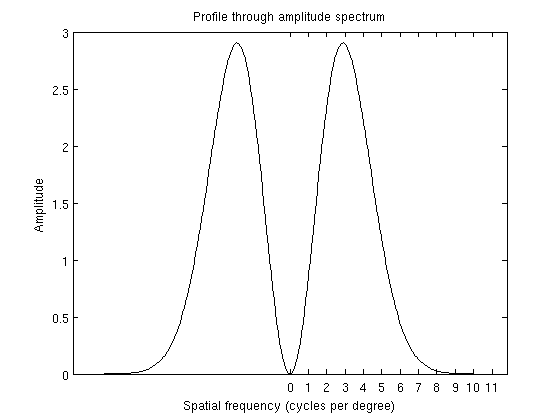
figure;
plot(-res/2:res/2-1,temp(res/2+1,:),'k-');
cpd = 0:20;
set(gca,'XTick',cpd*fov,'XTickLabel',cpd);
xlabel('Spatial frequency (cycles per degree)');
ylabel('Amplitude');
title('Profile through amplitude spectrum');