Example script illustrating how to fit the SOC model
Contents
Note
Add code to the MATLAB path
addpath(genpath(fullfile(pwd,'code')));
Load data
load('dataset03.mat','betamn','betase');
Load stimuli
load('stimuli.mat','images');
Perform stimulus pre-processing (Part 1)
stimulus = images(69+(1:99));
stimulus = cat(3,stimulus{:});
clear images;
temp = zeros(150,150,size(stimulus,3),'single');
for p=1:size(stimulus,3)
temp(:,:,p) = imresize(single(stimulus(:,:,p)),[150 150],'cubic');
end
stimulus = temp;
clear temp;
stimulus(stimulus < 0) = 0;
stimulus(stimulus > 254) = 254;
stimulus = stimulus/254 - 0.5;
stimulus = placematrix(zeros(180,180,size(stimulus,3),'single'),stimulus);
figure;
imagesc(stimulus(:,:,100));
axis image tight;
caxis([-.5 .5]);
colormap(gray);
colorbar;
title('Stimulus');
Perform stimulus pre-processing (Part 2)
stimulus = applymultiscalegaborfilters(reshape(stimulus,180*180,[])', ...
37.5*(180/150),-1,1,8,2,.01,2,0);
grid is 90 x 90, gbrsize is 14 (sd is 2.2486875016), indices is [1 3 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 35 37 39 41 43 45 47 49 51 53 55 57 59 61 63 65 67 69 71 73 75 77 79 81 83 85 87 89 91 93 95 97 99 101 103 105 107 109 111 113 115 117 119 121 123 125 127 129 131 133 135 137 139 141 143 145 147 149 151 153 155 157 159 161 163 165 167 169 171 173 175 177 179]
stimulus = sqrt(blob(stimulus.^2,2,2));
stimulusPOP = blob(stimulus,2,8)/8;
stimulusPOP = upsamplematrix(stimulusPOP,8,2,[],'nearest');
r = 1;
s = 0.5;
stimulus = stimulus.^r ./ (s.^r + stimulusPOP.^r);
clear stimulusPOP;
stimulus = blob(stimulus,2,8);
stimulus = permute(reshape(stimulus,9,99,8100),[2 3 1]);
figure;
mx = max(abs(stimulus(:)));
imagesc(reshape(stimulus(12,:,1),[90 90]));
axis image tight;
caxis([0 mx]);
colormap(gray);
colorbar;
title('Stimulus');
Prepare for model fitting
res = 90;
Ns = [.05 .1 .3 .5];
Cs = [.4 .7 .9 .95];
seeds = [];
for p=1:length(Ns)
for q=1:length(Cs)
seeds = cat(1,seeds,[(1+res)/2 (1+res)/2 res/4*sqrt(0.5) 10 Ns(p) Cs(q)]);
end
end
bounds = [1-res+1 1-res+1 0 -Inf 0 0;
2*res-1 2*res-1 Inf Inf Inf 1];
boundsFIX = bounds;
boundsFIX(1,5:6) = NaN;
[d,xx,yy] = makegaussian2d(res,2,2,2,2);
socfun = @(dd,wts,c) bsxfun(@minus,dd,c*(dd*wts)).^2 * wts;
gaufun = @(pp) vflatten(makegaussian2d(res,pp(1),pp(2),pp(3),pp(3),xx,yy,0,0)/(2*pi*pp(3)^2));
modelfun = @(pp,dd) pp(4)*(socfun(dd,gaufun(pp),restrictrange(pp(6),0,1)).^pp(5));
model = {{[] boundsFIX modelfun} ...
{@(ss) ss bounds @(ss) modelfun}};
optimoptions = {'Algorithm' 'levenberg-marquardt' 'Display' 'off'};
resampling = 0;
metric = @(a,b) calccod(a,b,[],[],0);
ix = 204;
opt = struct( ...
'stimulus', stimulus, ...
'data', betamn(ix,1:99)', ...
'model', {model}, ...
'seed', seeds, ...
'optimoptions', {optimoptions}, ...
'resampling', resampling, ...
'metric', metric);
opt
opt =
stimulus: [99x8100x9 single]
data: [99x1 double]
model: {{1x3 cell} {1x3 cell}}
seed: [16x6 double]
optimoptions: {'Algorithm' 'levenberg-marquardt' 'Display' 'off'}
resampling: 0
metric: @(a,b)calccod(a,b,[],[],0)
Fit the model
results = fitnonlinearmodel(opt);
*** fitnonlinearmodel: started at 28-Aug-2013 15:10:47. ***
*** fitnonlinearmodel: outputdir = , chunksize = 1, chunknum = 1
*** fitnonlinearmodel: processing voxel 1 (1 of 1). ***
starting resampling case 1 of 1.
trying seed 1 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.050 0.400 ].
the estimated parameters are [55.763 60.759 1.075 6.241 0.050 0.400 ].
for model 2 of 2, the seed is [55.763 60.759 1.075 6.241 0.050 0.400 ].
the estimated parameters are [53.152 57.367 1.289 6.004 0.105 0.193 ].
trying seed 2 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.050 0.700 ].
the estimated parameters are [56.422 61.855 1.082 6.701 0.050 0.700 ].
for model 2 of 2, the seed is [56.422 61.855 1.082 6.701 0.050 0.700 ].
the estimated parameters are [53.749 59.030 1.234 7.208 0.123 0.715 ].
trying seed 3 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.050 0.900 ].
the estimated parameters are [56.545 61.231 1.304 6.544 0.050 0.900 ].
for model 2 of 2, the seed is [56.545 61.231 1.304 6.544 0.050 0.900 ].
the estimated parameters are [54.061 57.207 1.392 9.173 0.168 0.862 ].
trying seed 4 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.050 0.950 ].
the estimated parameters are [80.008 87.283 4.416 9.646 0.050 0.950 ].
for model 2 of 2, the seed is [80.008 87.283 4.416 9.646 0.050 0.950 ].
the estimated parameters are [70.499 81.121 5.406 7.895 0.106 0.689 ].
trying seed 5 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.100 0.400 ].
the estimated parameters are [54.139 60.840 1.681 6.625 0.100 0.400 ].
for model 2 of 2, the seed is [54.139 60.840 1.681 6.625 0.100 0.400 ].
the estimated parameters are [54.105 61.161 1.635 7.906 0.147 0.669 ].
trying seed 6 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.100 0.700 ].
the estimated parameters are [73.527 76.438 5.492 7.006 0.100 0.700 ].
for model 2 of 2, the seed is [73.527 76.438 5.492 7.006 0.100 0.700 ].
the estimated parameters are [73.527 76.438 5.492 7.006 0.100 0.700 ].
trying seed 7 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.100 0.900 ].
the estimated parameters are [61.395 66.003 2.998 7.447 0.100 0.900 ].
for model 2 of 2, the seed is [61.395 66.003 2.998 7.447 0.100 0.900 ].
the estimated parameters are [56.215 61.618 2.810 9.365 0.201 0.849 ].
trying seed 8 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.100 0.950 ].
the estimated parameters are [60.598 70.078 3.031 7.260 0.100 0.950 ].
for model 2 of 2, the seed is [60.598 70.078 3.031 7.260 0.100 0.950 ].
the estimated parameters are [56.299 61.868 2.775 8.887 0.199 0.784 ].
trying seed 9 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.300 0.400 ].
the estimated parameters are [58.553 65.802 4.827 7.799 0.300 0.400 ].
for model 2 of 2, the seed is [58.553 65.802 4.827 7.799 0.300 0.400 ].
the estimated parameters are [57.411 63.666 3.724 9.288 0.222 0.816 ].
trying seed 10 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.300 0.700 ].
the estimated parameters are [60.161 62.370 5.223 9.567 0.300 0.700 ].
for model 2 of 2, the seed is [60.161 62.370 5.223 9.567 0.300 0.700 ].
the estimated parameters are [56.237 62.297 3.107 9.806 0.229 0.834 ].
trying seed 11 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.300 0.900 ].
the estimated parameters are [59.754 66.511 5.399 10.820 0.300 0.900 ].
for model 2 of 2, the seed is [59.754 66.511 5.399 10.820 0.300 0.900 ].
the estimated parameters are [60.425 66.786 5.401 10.466 0.284 0.904 ].
trying seed 12 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.300 0.950 ].
the estimated parameters are [60.237 66.872 5.742 10.710 0.300 0.950 ].
for model 2 of 2, the seed is [60.237 66.872 5.742 10.710 0.300 0.950 ].
the estimated parameters are [60.237 66.872 5.742 10.710 0.300 0.950 ].
trying seed 13 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.500 0.400 ].
the estimated parameters are [53.193 65.116 12.302 7.352 0.500 0.400 ].
for model 2 of 2, the seed is [53.193 65.116 12.302 7.352 0.500 0.400 ].
the estimated parameters are [56.460 61.532 3.112 10.141 0.241 0.840 ].
trying seed 14 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.500 0.700 ].
the estimated parameters are [56.418 59.072 11.814 10.450 0.500 0.700 ].
for model 2 of 2, the seed is [56.418 59.072 11.814 10.450 0.500 0.700 ].
the estimated parameters are [60.413 67.954 4.552 9.426 0.210 1.082 ].
trying seed 15 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.500 0.900 ].
the estimated parameters are [58.765 64.525 6.499 15.015 0.500 0.900 ].
for model 2 of 2, the seed is [58.765 64.525 6.499 15.015 0.500 0.900 ].
the estimated parameters are [59.142 64.366 6.525 14.717 0.484 0.894 ].
trying seed 16 of 16.
for model 1 of 2, the seed is [45.500 45.500 15.910 10.000 0.500 0.950 ].
the estimated parameters are [59.894 64.637 6.581 14.968 0.500 0.950 ].
for model 2 of 2, the seed is [59.894 64.637 6.581 14.968 0.500 0.950 ].
the estimated parameters are [60.064 65.534 6.221 14.066 0.442 1.020 ].
seed 3 was best. final estimated parameters are [54.061 57.207 1.392 9.173 0.168 0.862 ].
*** fitnonlinearmodel: voxel 1 (1 of 1) took 136.4 seconds. ***
*** fitnonlinearmodel: ended at 28-Aug-2013 15:13:04 (2.3 minutes). ***
Inspect the results
results.params
ans =
Columns 1 through 3
54.0608173443624 57.207123537548 1.39186915322788
Columns 4 through 6
9.17342790703929 0.168132144998856 0.861591769911232
results.trainperformance
ans =
95.1740363925398
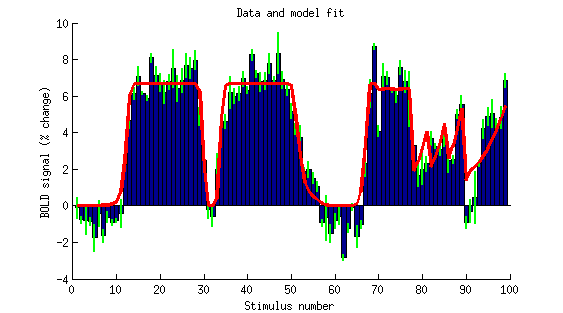
figure; setfigurepos([100 100 450 250]); hold on;
bar(betamn(ix,1:99),1);
errorbar2(1:99,betamn(ix,1:99),betase(ix,1:99),'v','g-','LineWidth',2);
modelfit = [];
for p=1:9
modelfit(:,p) = modelfun(results.params,stimulus(:,:,p));
end
plot(1:99,mean(modelfit,2),'r-','LineWidth',3);
ax = axis;
axis([0 100 ax(3:4)]);
xlabel('Stimulus number');
ylabel('BOLD signal (% change)');
title('Data and model fit');