Example 2: Analyze different splits of runs in a session
Contents
Download dataset (if necessary) and add GLMdenoise to the MATLAB path
setup;
Load in the data
load('exampledataset.mat');
whos
Name Size Bytes Class Attributes
data 1x10 173671520 cell
design 1x10 9600 cell
stimdur 1x1 8 double
tr 1x1 8 double
Split session into two halves and analyze each half separately
splits = {[1:2:10] [2:2:10]};
clear resultsREGULAR resultsDENOISE;
for p=1:length(splits)
ix = splits{p};
resultsREGULAR(p) = GLMdenoisedata(design(ix),data(ix),stimdur,tr, ...
[],[],struct('numpcstotry',0), ...
sprintf('example2figures_REGULAR_split%d',p));
resultsDENOISE(p) = GLMdenoisedata(design(ix),data(ix),stimdur,tr, ...
[],[],[], ...
sprintf('example2figures_DENOISE_split%d',p));
end
*** GLMdenoisedata: generating sanity-check figures. ***
*** GLMdenoisedata: performing full fit to estimate global HRF. ***
fitting model...done.
preparing output...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation to determine R^2 values. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: determining noise pool. ***
*** GLMdenoisedata: calculating noise regressors. ***
*** GLMdenoisedata: selected number of PCs is 0. ***
*** GLMdenoisedata: fitting final model (no denoising, for comparison purposes). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: fitting final model (with denoising). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: calculating denoised data and PC weights. ***
*** GLMdenoisedata: converting to percent BOLD change. ***
*** GLMdenoisedata: generating figures. ***
*** GLMdenoisedata: generating sanity-check figures. ***
*** GLMdenoisedata: performing full fit to estimate global HRF. ***
fitting model...done.
preparing output...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation to determine R^2 values. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: determining noise pool. ***
*** GLMdenoisedata: calculating noise regressors. ***
*** GLMdenoisedata: performing cross-validation with 1 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 2 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 3 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 4 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 5 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 6 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 7 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 8 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 9 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 10 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 11 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 12 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 13 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 14 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 15 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 16 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 17 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 18 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 19 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 20 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: selected number of PCs is 5. ***
*** GLMdenoisedata: fitting final model (no denoising, for comparison purposes). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: fitting final model (with denoising). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: calculating denoised data and PC weights. ***
*** GLMdenoisedata: converting to percent BOLD change. ***
*** GLMdenoisedata: generating figures. ***
*** GLMdenoisedata: generating sanity-check figures. ***
*** GLMdenoisedata: performing full fit to estimate global HRF. ***
fitting model...done.
preparing output...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation to determine R^2 values. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: determining noise pool. ***
*** GLMdenoisedata: calculating noise regressors. ***
*** GLMdenoisedata: selected number of PCs is 0. ***
*** GLMdenoisedata: fitting final model (no denoising, for comparison purposes). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: fitting final model (with denoising). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: calculating denoised data and PC weights. ***
*** GLMdenoisedata: converting to percent BOLD change. ***
*** GLMdenoisedata: generating figures. ***
*** GLMdenoisedata: generating sanity-check figures. ***
*** GLMdenoisedata: performing full fit to estimate global HRF. ***
fitting model...done.
preparing output...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation to determine R^2 values. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: determining noise pool. ***
*** GLMdenoisedata: calculating noise regressors. ***
*** GLMdenoisedata: performing cross-validation with 1 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 2 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 3 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 4 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 5 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 6 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 7 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 8 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 9 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 10 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 11 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 12 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 13 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 14 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 15 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 16 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 17 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 18 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 19 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 20 PCs. ***
cross-validating model.....done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: selected number of PCs is 5. ***
*** GLMdenoisedata: fitting final model (no denoising, for comparison purposes). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: fitting final model (with denoising). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: calculating denoised data and PC weights. ***
*** GLMdenoisedata: converting to percent BOLD change. ***
*** GLMdenoisedata: generating figures. ***
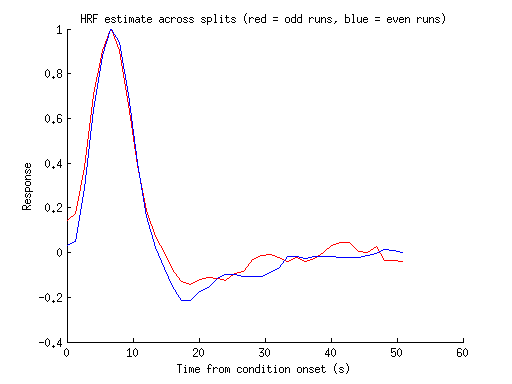
figure; hold on;
hrf1 = resultsREGULAR(1).modelmd{1};
hrf2 = resultsREGULAR(2).modelmd{1};
plot(0:tr:tr*(length(hrf1)-1),hrf1,'r-');
plot(0:tr:tr*(length(hrf2)-1),hrf2,'b-');
xlabel('Time from condition onset (s)');
ylabel('Response');
title('HRF estimate across splits (red = odd runs, blue = even runs)');
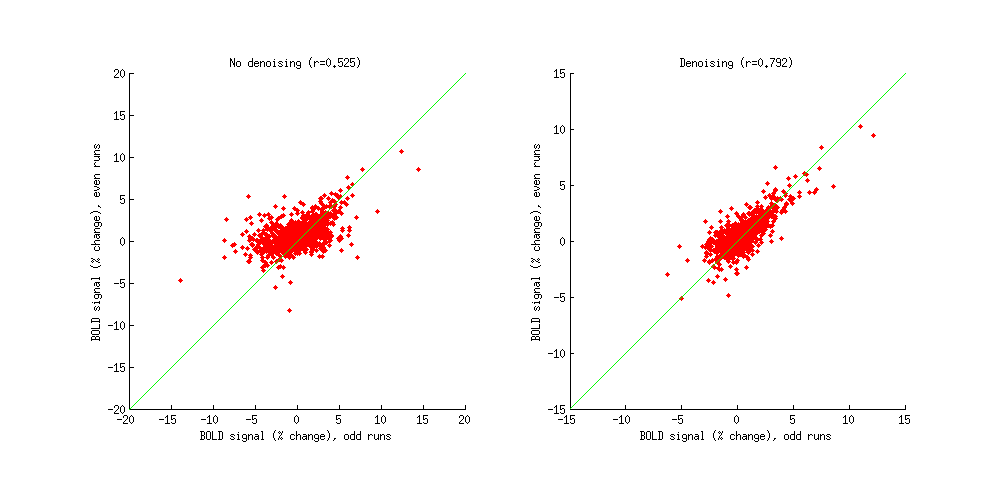
ix = find(resultsREGULAR(1).R2 > 10 & resultsREGULAR(1).meanvol > 500);
beta1REGULAR = subscript(squish(resultsREGULAR(1).modelmd{2},3),{ix 10});
beta2REGULAR = subscript(squish(resultsREGULAR(2).modelmd{2},3),{ix 10});
beta1DENOISE = subscript(squish(resultsDENOISE(1).modelmd{2},3),{ix 10});
beta2DENOISE = subscript(squish(resultsDENOISE(2).modelmd{2},3),{ix 10});
figure;
set(gcf,'Units','points','Position',[100 100 800 400]);
subplot(1,2,1); hold on;
scatter(beta1REGULAR,beta2REGULAR,'r.');
axissquarify;
xlabel('BOLD signal (% change), odd runs');
ylabel('BOLD signal (% change), even runs');
title(sprintf('No denoising (r=%.3f)',corr(beta1REGULAR,beta2REGULAR)));
subplot(1,2,2); hold on;
scatter(beta1DENOISE,beta2DENOISE,'r.');
axissquarify;
xlabel('BOLD signal (% change), odd runs');
ylabel('BOLD signal (% change), even runs');
title(sprintf('Denoising (r=%.3f)',corr(beta1DENOISE,beta2DENOISE)));
Split session into individual runs and analyze each run separately
splits = num2cell(1:10);
hrf = getcanonicalhrf(stimdur,tr)';
clear resultsIND;
for p=1:length(splits)
ix = splits{p};
resultsIND(p) = GLMestimatemodel(design(ix),data(ix),stimdur,tr,'assume',hrf,0);
end
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
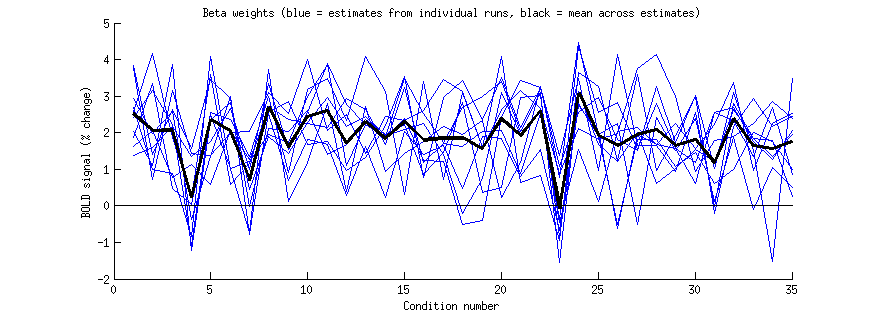
xx = 46; yy = 24; zz = 1;
figure; hold on;
set(gcf,'Units','points','Position',[100 100 700 250]);
betas = [];
for p=1:length(resultsIND)
betas(p,:) = flatten(resultsIND(p).modelmd{2}(xx,yy,zz,:));
plot(betas(p,:),'b-');
end
plot(mean(betas,1),'k-','LineWidth',3);
straightline(0,'h','k-');
xlabel('Condition number');
ylabel('BOLD signal (% change)');
title('Beta weights (blue = estimates from individual runs, black = mean across estimates)');