Example 1: Run GLMdenoise on an example dataset
Contents
Download dataset (if necessary) and add GLMdenoise to the MATLAB path
setup;
Load in the data
load('exampledataset.mat');
whos
Name Size Bytes Class Attributes
data 1x10 173671520 cell
design 1x10 9600 cell
stimdur 1x1 8 double
tr 1x1 8 double
Inspect the data
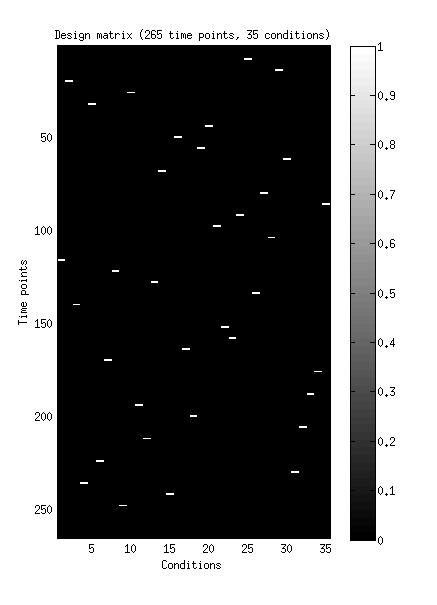
figure;
set(gcf,'Units','points','Position',[100 100 350 500]);
imagesc(design{1});
colormap(gray);
colorbar;
xlabel('Conditions');
ylabel('Time points');
title(sprintf('Design matrix (%d time points, %d conditions)',size(design{1},1),size(design{1},2)));
figure;
imagesc(makeimagestack(data{1}(:,:,:,1)));
colormap(gray);
axis equal tight off;
colorbar;
title('fMRI data (first volume)');
fprintf('There are %d runs in total.\n',length(design));
fprintf('The dimensions of the data for the first run are %s.\n',mat2str(size(data{1})));
fprintf('The stimulus duration is %.6f seconds.\n',stimdur);
fprintf('The sampling rate (TR) is %.6f seconds.\n',tr);
There are 10 runs in total.
The dimensions of the data for the first run are [64 64 4 265].
The stimulus duration is 3.000000 seconds.
The sampling rate (TR) is 1.337702 seconds.
Run GLMdenoise
[results,denoiseddata] = GLMdenoisedata(design,data,stimdur,tr,[],[],[],'example1figures');
*** GLMdenoisedata: performing full fit to estimate global HRF. ***
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation to determine R^2 values. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
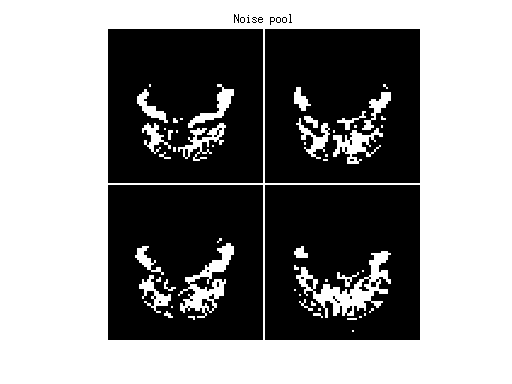
*** GLMdenoisedata: determining noise pool. ***
*** GLMdenoisedata: calculating global noise regressors. ***
*** GLMdenoisedata: performing cross-validation with 1 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 2 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 3 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 4 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 5 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 6 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 7 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 8 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 9 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 10 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 11 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 12 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 13 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 14 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 15 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 16 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 17 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 18 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 19 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation with 20 PCs. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
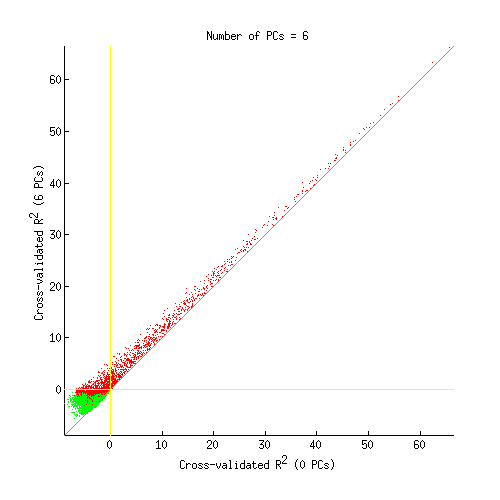
*** GLMdenoisedata: selected number of PCs is 6. ***
*** GLMdenoisedata: fitting final model (no denoising, for comparison purposes). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: fitting final model (with denoising). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: calculating denoised data and PC weights. ***
*** GLMdenoisedata: converting to percent BOLD change. ***
*** GLMdenoisedata: generating figures. ***
[resultsALT,denoiseddataALT] = GLMdenoisedata(design,data,stimdur,tr,[],[],struct('numpcstotry',0),'example1figuresALT');
*** GLMdenoisedata: performing full fit to estimate global HRF. ***
fitting model...done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: performing cross-validation to determine R^2 values. ***
cross-validating model..........done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: determining noise pool. ***
*** GLMdenoisedata: calculating global noise regressors. ***
*** GLMdenoisedata: selected number of PCs is 0. ***
*** GLMdenoisedata: fitting final model (no denoising, for comparison purposes). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: fitting final model (with denoising). ***
bootstrapping model....................done.
preparing output...done.
computing model fits...done.
computing R^2...done.
computing SNR...done.
*** GLMdenoisedata: calculating denoised data and PC weights. ***
*** GLMdenoisedata: converting to percent BOLD change. ***
*** GLMdenoisedata: generating figures. ***
Inspect figures
figure;
imagesc(imread('example1figures/MeanVolume.png'),[0 255]);
colormap(gray);
axis equal tight off;
title('Mean volume');

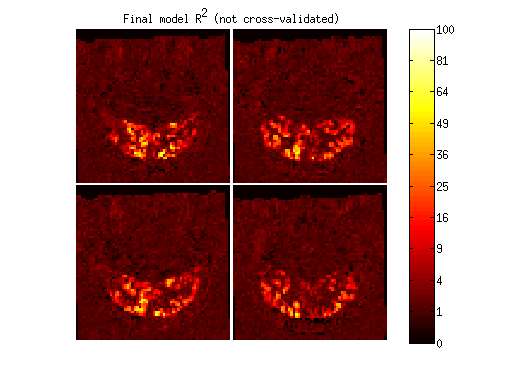
figure;
imagesc(imread('example1figures/FinalModel.png'),[0 255]);
colormap(hot);
axis equal tight off;
cb = colorbar;
set(cb,'YTick',linspace(0,255,11),'YTickLabel',round((0:.1:1).^2 * 100));
title('Final model R^2 (not cross-validated)');
figure;
imagesc(imread('example1figures/NoisePool.png'),[0 255]);
colormap(gray);
axis equal tight off;
title('Noise pool');
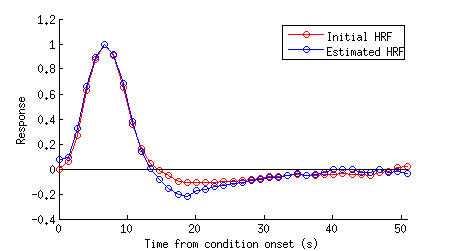
figure;
imageactual('example1figures/HRF.png');
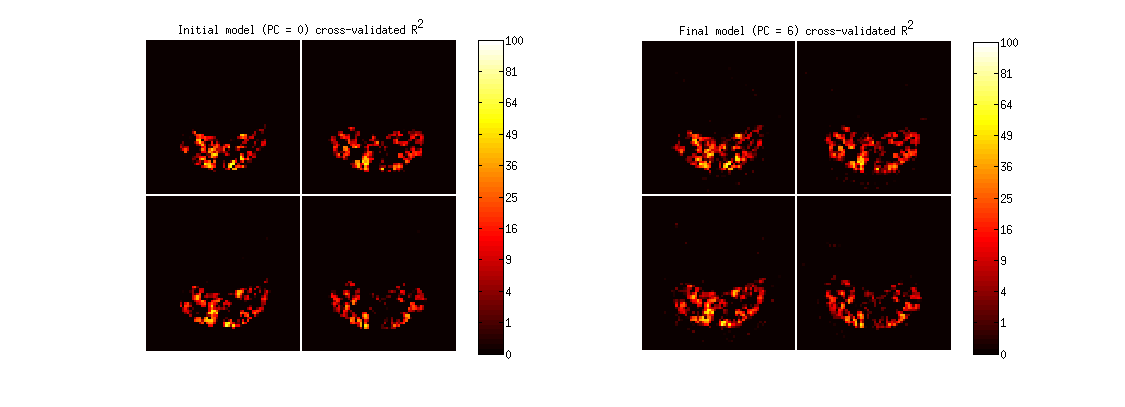
figure;
set(gcf,'Units','points','Position',[100 100 900 325]);
subplot(1,2,1);
imagesc(imread('example1figures/PCcrossvalidation00.png'),[0 255]);
colormap(hot);
axis equal tight off;
cb = colorbar;
set(cb,'YTick',linspace(0,255,11),'YTickLabel',round((0:.1:1).^2 * 100));
title('Initial model (PC = 0) cross-validated R^2');
subplot(1,2,2);
imagesc(imread(sprintf('example1figures/PCcrossvalidation%02d.png',results.pcnum)));
colormap(hot);
axis equal tight off;
cb = colorbar;
set(cb,'YTick',linspace(0,255,11),'YTickLabel',round((0:.1:1).^2 * 100));
title(sprintf('Final model (PC = %d) cross-validated R^2',results.pcnum));
figure;
imageactual(sprintf('example1figures/PCscatter%02d.png',results.pcnum));
figure;
imageactual('example1figures/PCselection.png');

Inspect outputs
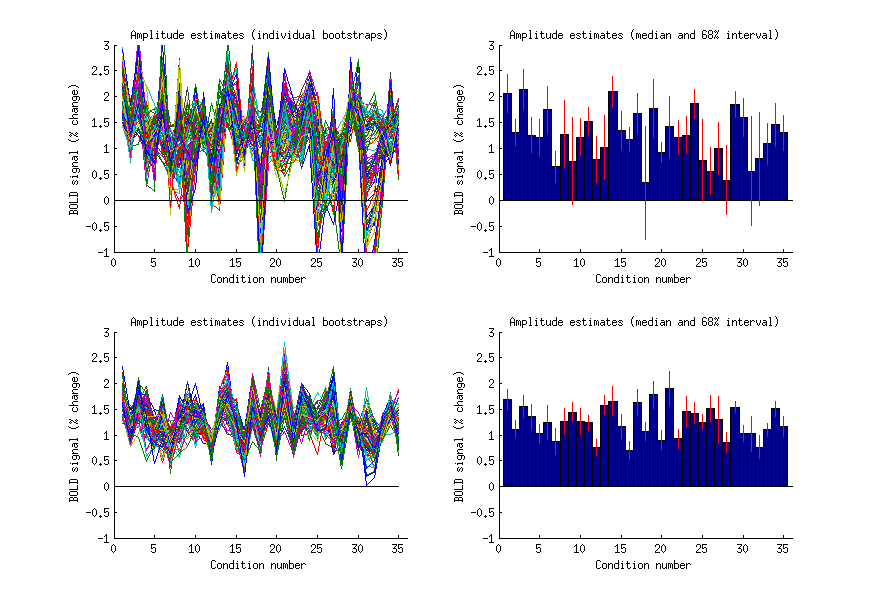
ix = find(results.pcR2(:,:,:,1) > 0 & results.pcR2(:,:,:,1) < 5);
improvement = results.pcR2(:,:,:,1+results.pcnum) - results.pcR2(:,:,:,1);
[mm,ii] = max(improvement(ix));
ix = ix(ii);
[xx,yy,zz] = ind2sub(results.inputs.datasize{1}(1:3),ix);
figure;
set(gcf,'Units','points','Position',[100 100 700 500]);
for p=1:2
if p==1
ampboots = squeeze(resultsALT.models{2}(xx,yy,zz,:,:));
amp = flatten(resultsALT.modelmd{2}(xx,yy,zz,:));
ampse = flatten(resultsALT.modelse{2}(xx,yy,zz,:));
else
ampboots = squeeze(results.models{2}(xx,yy,zz,:,:));
amp = flatten(results.modelmd{2}(xx,yy,zz,:));
ampse = flatten(results.modelse{2}(xx,yy,zz,:));
end
n = length(amp);
subplot(2,2,(p-1)*2 + 1); hold on;
plot(ampboots);
straightline(0,'h','k-');
xlabel('Condition number');
ylabel('BOLD signal (% change)');
title('Amplitude estimates (individual bootstraps)');
subplot(2,2,(p-1)*2 + 2); hold on;
bar(1:length(amp),amp,1);
errorbar2(1:length(amp),amp,ampse,'v','r-');
if p==1
ax = axis; axis([0 n+1 ax(3:4)]); ax = axis;
end
xlabel('Condition number');
ylabel('BOLD signal (% change)');
title('Amplitude estimates (median and 68% interval)');
end
for p=1:2
subplot(2,2,(p-1)*2 + 1); axis(ax);
subplot(2,2,(p-1)*2 + 2); axis(ax);
end
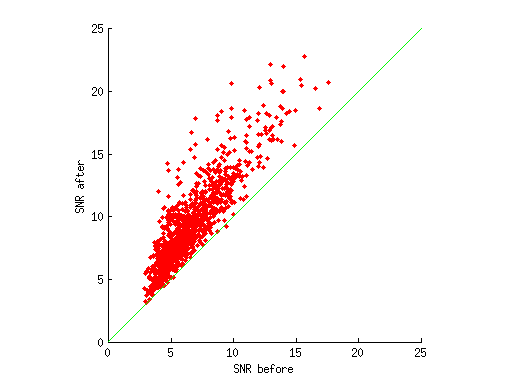
ok = results.pcR2(:,:,:,1) > 0;
signal = mean([results.signal(ok) resultsALT.signal(ok)],2);
snr1 = signal ./ resultsALT.noise(ok);
snr2 = signal ./ results.noise(ok);
figure; hold on;
scatter(snr1,snr2,'r.');
ax = axis;
mx = max(ax(3:4));
axissquarify;
axis([0 mx 0 mx]);
xlabel('SNR before');
ylabel('SNR after');
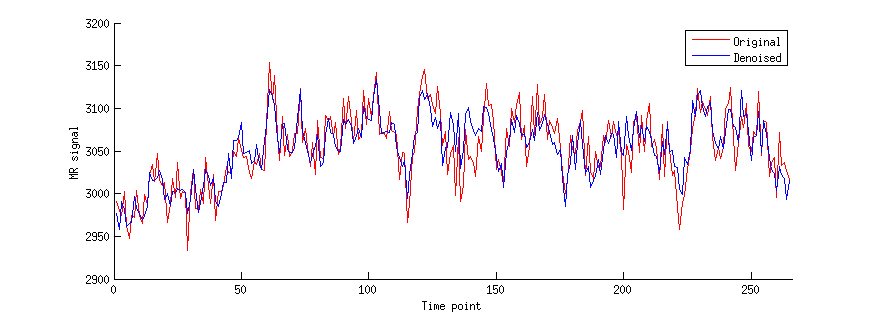
figure; hold on;
set(gcf,'Units','points','Position',[100 100 700 250]);
data1 = flatten(data{1}(xx,yy,zz,:));
data2 = flatten(denoiseddata{1}(xx,yy,zz,:));
n = length(data1);
h1 = plot(data1,'r-');
h2 = plot(data2,'b-');
ax = axis; axis([0 n+1 ax(3:4)]);
legend([h1 h2],{'Original' 'Denoised'});
xlabel('Time point');
ylabel('MR signal');