Generate figures illustrating the DNB results
Contents
Download the DNB results (if necessary)
DNBdownloadresults;
dnbdir = absolutepath(strrep(which('DNBrun'),'DNBrun.m',''));
mkdirquiet(fullfile(dnbdir,'figures'));
mkdirquiet(fullfile(dnbdir,'figures','scatter'));
Downloading results.mat (please be patient).
Downloading is done!
Downloading figures.tar (please be patient).
Downloading is done!
Load in a .mat file that contains the DNB results
load(fullfile(dnbdir,'DNBresults','results.mat'));
Check the workspace
whos
Name Size Bytes Class Attributes
allR2 21x11 92086208 cell
brainmasks 1x21 2331648 cell
denoisemethods 1x11 1522 cell
denoisenames 1x11 1550 cell
dnbdir 1x14 28 char
meanvols 1x21 9319536 cell
voxelselections 1x21 2331648 cell
Write out spatial maps of cross-validated R^2 values
for p=1:size(allR2,1)
fprintf('writing cross-validated R^2 maps for dataset %d...',p);
for q=1:size(allR2,2)
if ~isempty(allR2{p,q})
file0 = sprintf(fullfile(dnbdir,'figures','dataset%02d_%s.png'),p,denoisemethods{q});
imwrite(uint8(255*makeimagestack(signedarraypower(allR2{p,q}/100,0.5),[0 1])),hot(256),file0);
end
end
fprintf('done.\n');
end
writing cross-validated R^2 maps for dataset 1...done.
writing cross-validated R^2 maps for dataset 2...done.
writing cross-validated R^2 maps for dataset 3...done.
writing cross-validated R^2 maps for dataset 4...done.
writing cross-validated R^2 maps for dataset 5...done.
writing cross-validated R^2 maps for dataset 6...done.
writing cross-validated R^2 maps for dataset 7...done.
writing cross-validated R^2 maps for dataset 8...done.
writing cross-validated R^2 maps for dataset 9...done.
writing cross-validated R^2 maps for dataset 10...done.
writing cross-validated R^2 maps for dataset 11...done.
writing cross-validated R^2 maps for dataset 12...done.
writing cross-validated R^2 maps for dataset 13...done.
writing cross-validated R^2 maps for dataset 14...done.
writing cross-validated R^2 maps for dataset 15...done.
writing cross-validated R^2 maps for dataset 16...done.
writing cross-validated R^2 maps for dataset 17...done.
writing cross-validated R^2 maps for dataset 18...done.
writing cross-validated R^2 maps for dataset 19...done.
writing cross-validated R^2 maps for dataset 20...done.
writing cross-validated R^2 maps for dataset 21...done.
Inspect a cross-validated R^2 map
figure;
imageactual(fullfile(dnbdir,'figures','dataset01_GLMdenoise.png'));
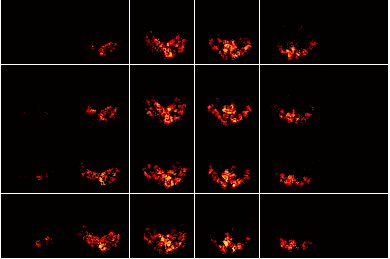
Write out spatial maps that provide additional information
for p=1:length(meanvols)
fprintf('writing additional maps for dataset %d...',p);
file0 = sprintf(fullfile(dnbdir,'figures','dataset%02d_meanvol.png'),p);
imwrite(uint8(255*makeimagestack(meanvols{p},1)),gray(256),file0);
file0 = sprintf(fullfile(dnbdir,'figures','dataset%02d_voxelselection.png'),p);
imwrite(uint8(255*makeimagestack(voxelselections{p},[0 1])),gray(256),file0);
file0 = sprintf(fullfile(dnbdir,'figures','dataset%02d_brainmask.png'),p);
imwrite(uint8(255*makeimagestack(brainmasks{p},[0 1])),gray(256),file0);
fprintf('done.\n');
end
writing additional maps for dataset 1...done.
writing additional maps for dataset 2...done.
writing additional maps for dataset 3...done.
writing additional maps for dataset 4...done.
writing additional maps for dataset 5...done.
writing additional maps for dataset 6...done.
writing additional maps for dataset 7...done.
writing additional maps for dataset 8...done.
writing additional maps for dataset 9...done.
writing additional maps for dataset 10...done.
writing additional maps for dataset 11...done.
writing additional maps for dataset 12...done.
writing additional maps for dataset 13...done.
writing additional maps for dataset 14...done.
writing additional maps for dataset 15...done.
writing additional maps for dataset 16...done.
writing additional maps for dataset 17...done.
writing additional maps for dataset 18...done.
writing additional maps for dataset 19...done.
writing additional maps for dataset 20...done.
writing additional maps for dataset 21...done.
Inspect these additional maps
figure;
imageactual(fullfile(dnbdir,'figures','dataset01_meanvol.png'));
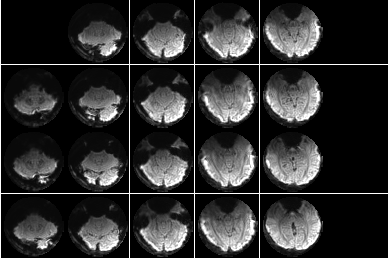
figure;
imageactual(fullfile(dnbdir,'figures','dataset01_voxelselection.png'));
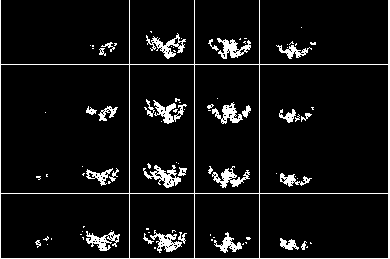
figure;
imageactual(fullfile(dnbdir,'figures','dataset01_brainmask.png'));
For each denoising method, calculate the median cross-validated R^2 value achieved on each dataset
summary = zeros([size(allR2) 3]);
for p=1:size(allR2,1)
fprintf('calculating median R^2 values for dataset %d...',p);
for q=1:size(allR2,2)
if isempty(allR2{p,q})
summary(p,q,1:3) = NaN;
else
[d,d,summary(p,q,1:3)] = calcmdsepct(allR2{p,q}(voxelselections{p}));
end
end
fprintf('done.\n');
end
calculating median R^2 values for dataset 1...done.
calculating median R^2 values for dataset 2...done.
calculating median R^2 values for dataset 3...done.
calculating median R^2 values for dataset 4...done.
calculating median R^2 values for dataset 5...done.
calculating median R^2 values for dataset 6...done.
calculating median R^2 values for dataset 7...done.
calculating median R^2 values for dataset 8...done.
calculating median R^2 values for dataset 9...done.
calculating median R^2 values for dataset 10...done.
calculating median R^2 values for dataset 11...done.
calculating median R^2 values for dataset 12...done.
calculating median R^2 values for dataset 13...done.
calculating median R^2 values for dataset 14...done.
calculating median R^2 values for dataset 15...done.
calculating median R^2 values for dataset 16...done.
calculating median R^2 values for dataset 17...done.
calculating median R^2 values for dataset 18...done.
calculating median R^2 values for dataset 19...done.
calculating median R^2 values for dataset 20...done.
calculating median R^2 values for dataset 21...done.
Visualize a summary of the cross-validation results on a bar chart
ixs = {1:7 8:13 14:21};
for p=1:length(ixs)
figure; set(gcf,'Visible','off','PaperPositionMode','auto');
setfigurepos([100 100 900 275]); hold on;
h = bar(summary(ixs{p},:,1),1);
colormap(jet);
for mm=1:length(h)
tempxd = get(get(h(mm),'Children'),'XData');
errorbar2(mean(tempxd([1 3],:),1),summary(ixs{p},mm,1)', ...
squish(summary(ixs{p},mm,2:3),2)','v','r-','LineWidth',1);
end
set(gca,'XTick',1:length(ixs{p}),'XTickLabel',ixs{p});
xlabel('Dataset');
ylabel('Cross-validated R^2');
legend(h,denoisenames,'Location','EastOutside');
filename = sprintf(fullfile(dnbdir,'figures','summary%d.png'),p);
print('-dpng','-r72',filename);
close;
figure;
imageactual(filename);
end
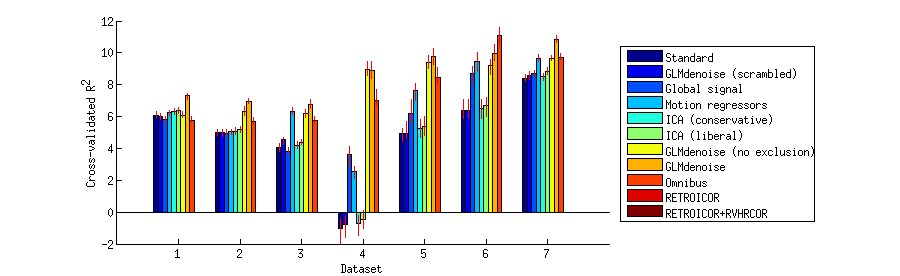
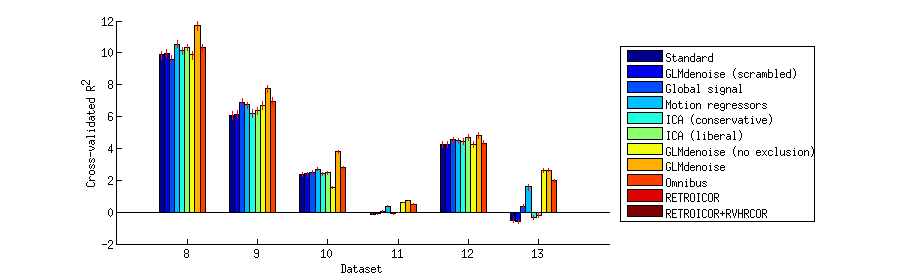
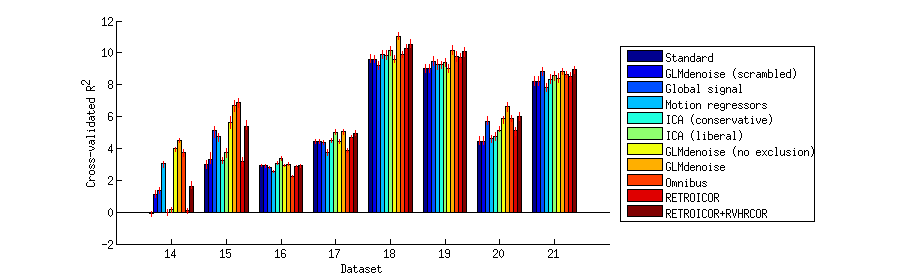
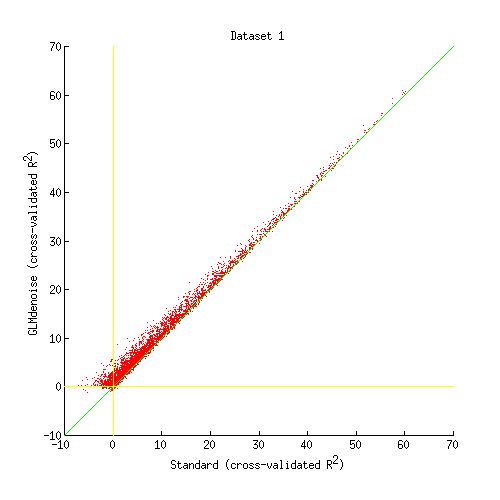
Write out scatter plots that show a detailed comparison of each pair of denoising methods on each dataset
for p=1:size(allR2,1)
fprintf('writing scatter plots for dataset %d...',p);
for d1=1:size(allR2,2)
for d2=1:size(allR2,2)
if isempty(allR2{p,d1}) || isempty(allR2{p,d2}) || d1==d2
continue;
end
figure; set(gcf,'Visible','off','PaperPositionMode','auto');
setfigurepos([100 100 500 500]); hold on;
scatter(allR2{p,d1}(voxelselections{p}), ...
allR2{p,d2}(voxelselections{p}),25,'r.');
ax = axis;
mn = min(ax([1 3]));
mx = max(ax([2 4]));
axis([mn mx mn mx]);
axissquarify;
axis([mn mx mn mx]);
straightline(0,'h','y-');
straightline(0,'v','y-');
xlabel(sprintf('%s (cross-validated R^2)',denoisenames{d1}));
ylabel(sprintf('%s (cross-validated R^2)',denoisenames{d2}));
title(sprintf('Dataset %d',p));
filename = sprintf(fullfile(dnbdir,'figures','scatter','scatter_%s_vs_%s_dataset%02d.png'), ...
denoisemethods{d1},denoisemethods{d2},p);
print('-dpng','-r72',filename);
close;
end
end
fprintf('done.\n');
end
writing scatter plots for dataset 1...done.
writing scatter plots for dataset 2...done.
writing scatter plots for dataset 3...done.
writing scatter plots for dataset 4...done.
writing scatter plots for dataset 5...done.
writing scatter plots for dataset 6...done.
writing scatter plots for dataset 7...done.
writing scatter plots for dataset 8...done.
writing scatter plots for dataset 9...done.
writing scatter plots for dataset 10...done.
writing scatter plots for dataset 11...done.
writing scatter plots for dataset 12...done.
writing scatter plots for dataset 13...done.
writing scatter plots for dataset 14...done.
writing scatter plots for dataset 15...done.
writing scatter plots for dataset 16...done.
writing scatter plots for dataset 17...done.
writing scatter plots for dataset 18...done.
writing scatter plots for dataset 19...done.
writing scatter plots for dataset 20...done.
writing scatter plots for dataset 21...done.
Inspect one of these scatter plots
figure;
imageactual(fullfile(dnbdir,'figures','scatter','scatter_GLMstandard_vs_GLMdenoise_dataset01.png'));